Dates, time, and place January 26 – 30, 2015, noon to 1:00 PM in room WC130 on the FIU Modesto A. Maidique Campus (MMC), simultaneously broadcast to room MSB105 on the Biscayne Bay Campus (BBC). [Wednesday, January 28 the presentation will be broadcast from BBC to MMC.] A 30-minute period will be available for discussion following each presentation.

Dr. Steven Henikoff
Fred Hutchinson Cancer Research Center/Howard Hughes Medical Institute/University of Washington
Member of the National Academy of Sciences and Fellow of the American Association for the Advancement of Science.
Editor-in-Chief of the journal “Epigenetics & Chromatin”
Link to Dr. Henikoff laboratory web site
Link to Dr. Henikoff National Academy of Sciences profile
BACKGROUND
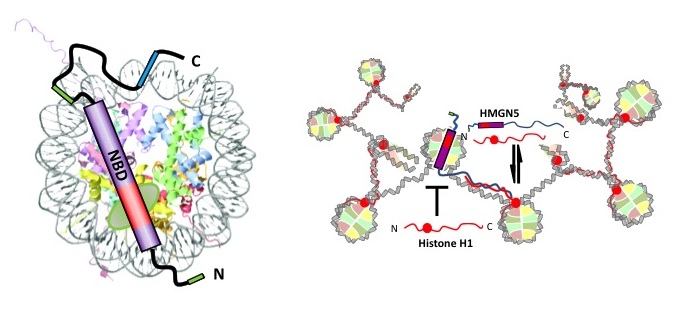
For more than a century histone proteins, composed of a polymer known as chromatin, have been studied for their role in the physical organization of chromosomes. More recently interest in this complex has been greatly rekindled by the recognition of chromatin’s role in gene regulation, including acquired traits that are inherited without involving changes in the DNA sequence. In other words, chromatin plays a central role in the configuration and propagation of epigenetic information across generations. The rapid increases in “omic” data and analytical methods have provided new approaches for understanding the interaction between genetic and epigenetic information, adding a layer of complexity in DNA regulation that, while still poorly understood, will clearly be important for understanding how cells adapt to changes in external conditions.
The research of Dr. Steve Henikoff (BS University of Chicago, PhD Harvard University) and his team seeks to understand this elusive relationship between chromatin and epigenetic inheritance. Their work has applied pioneering research approaches combining molecular biology, genetics, evolution and bioinformatics to study the different mechanisms potentially encoding epigenetic information (i.e., histone variants and their chemical modifications, DNA methylation, transcription factor regulatory networks, among others). Among these works, the characterization of the mechanisms governing centromeric chromatin structure and evolution are specially groundbreaking, challenging the classic notion stating that our whole genome, our chromosomes, are filled up with nucleosomes and they’re all octamers. In recent years they have expanded their scope from chromatin structure, dynamics and evolution to interactions with other components of the epigenome including nucleosome remodelers, transcription factors and RNA polymerase II*.
LECTURE TOPICS
Monday, Jan 26: Chromatin dynamics.
Tuesday, Jan 27: Transcription and chromatin.
Wednesday, Jan 28: Centromeric chromatin.
Thursday, Jan 29: Chromatin and cancer.
Friday, Jan 30: Chromatin and centromere evolution.
*From the Henikoff lab web site.