Our last work has been just published today on Peer J!
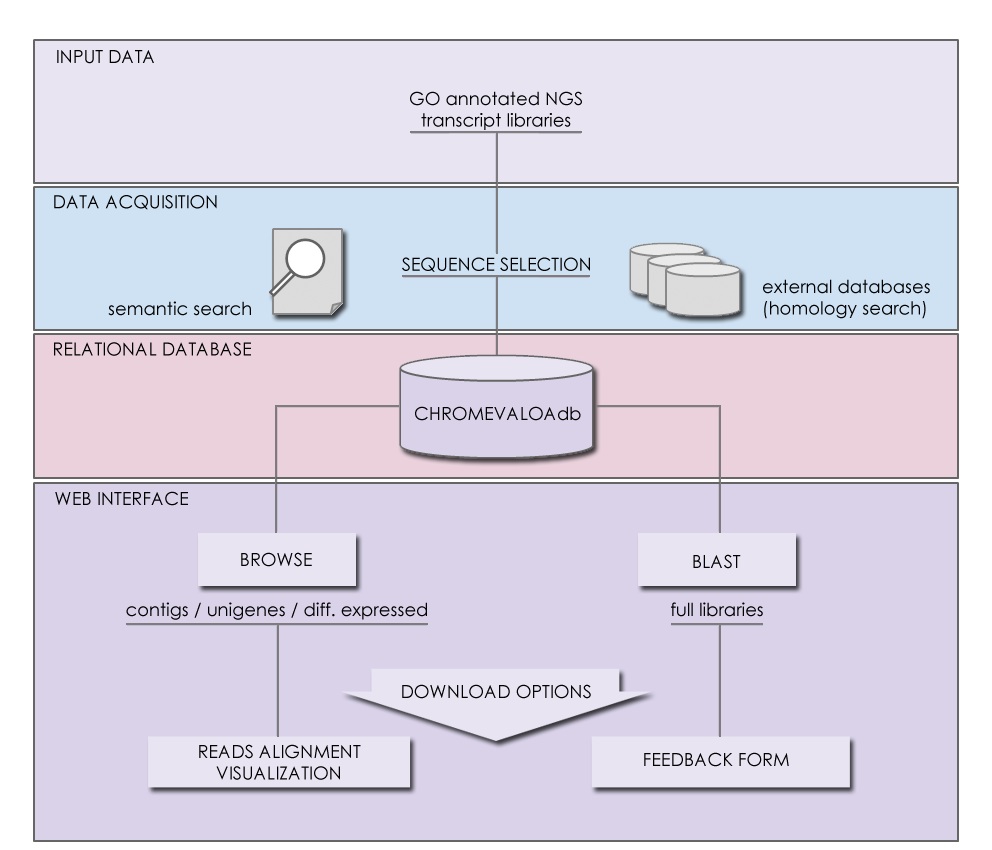
Posted by Chromevol | Bioinformatics, DNA, Marine Biology, Omics, Red Tides, Research, Toxicology | No CommentsVictoria (Vicky) Suarez-Ulloa, a Graduate Student at Chromevol, has led our last paper entitled “Unbiased high-throughput characterization of mussel transcriptomic responses to sublethal concentrations of the biotoxin okadaic acid”, published today in the journal Peer J. In this work we have collaborated with researchers from UK and Spain characterizing the genes involved in responses to the effects of marine biotoxins. In the pictures below you can see the now traditional “hanging ceremony” of the paper reprint below the corresponding poster (presented at the Gordon Conference in Ecological and Evolutionary Genomics last summer) in the hallway near our lab. Vicky was fortunate enough to count with the help and indications of three gentleman during this operation.
Congrats!!!