Email: chapagap@fiu.edu
Research Topics
We use molecular modeling and molecular dynamics simulations to investigate a variety of biological systems. With the access to powerful GPU computing resources, we can simulate large systems for microsecond timescales. The topics of our computational investigations are quite diverse. Some of the major projects pursued in our lab include the following:
Structure and dynamics of the Spike protein of SARS-CoV-2 variants
The Covid-19 pandemic caused by the SARS-CoV-2 coronavirus infections started in late 2019 is responsible for hundreds of millions of cases worldwide and millions of fatalities. The combined efforts of the researchers have achieved much needed success for both therapeutics and vaccines against the SARS-CoV-2 infections. Though current vaccines offer protection against known variants of concern, the virus continues to mutate as it spreads. Our research focuses on the effects of the SARS-CoV-2 mutations on the structure of the Spike protein leading to changes in the receptor binding as well as immune evasion. We have computationally investigated the structural dynamics of the receptor binding interface of various strains, including the Delta variant, and studied how these mutations allow the visddrus strains to function differently. We are also involved in a team effort to create an AI-driven pipeline utilizing molecular mimicry of antibody-antigen interactions.
This project is supported by the NSF.
Membrane interactions and conformational changes in the Filovirus protein VP40
Ebolavirus (EBOV) is a negative-sense RNA virus that causes severe hemorrhagic fever in humans with a high fatality rate. Although EBOV genome encodes only eight proteins, conformational changes and alternate structures allow these proteins, particularly the matrix protein VP40, to perform multiple functions. Viral budding involves lipid interactions of the virus’ matrix protein VP40 at the lower leaflet of the host’s cell membrane. Interestingly, VP40 alone is able to form authentic-looking virus like particles (VLPs) that bud from the host cell. We investigate how VP40 interacts with lipids and associates with and assembles at the host’s membrane surface that ultimately leads to the formation of long filamentous, pleomorphic virus particles. In collaboration with the Stahelin lab (Purdue) and Weist lab (Notre-Dame), we are focusing on various aspects of the VP40-membrane interactions, including protein conformational changes, roles of specific lipid types in membrane association, and the effects of the mutations found in disease outbreaks. This project is supported by the NIH.
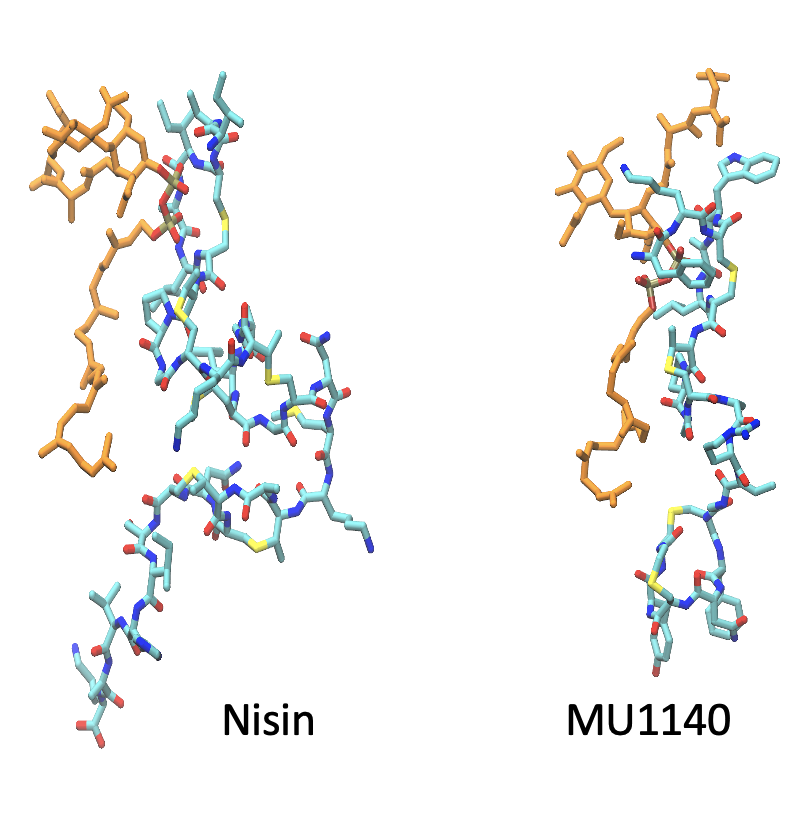
Computer-aided design of novel antimicrobial lantibiotic peptides
There has been an alarming rise in antibacterial resistance infections in recent years. Without a major advancement in new class of antibiotics soon, we will be facing a post-antibiotic era in which frequent infections and minor injuries can cost numerous lives and medical costs. Lantibiotics or lanthipeptides are promising candidates to mitigate the threat of antibiotic resistance. Lantibiotics are ribosomally synthesized peptides that contain post-translationally modified unusual amino acids including lanthionine. Lantibiotics commonly target lipid II which is an essential component for creating the peptidoglycan layer of bacterial cell wall but some also disrupt membrane or form membrane pores. We use molecular modeling and computer simulations to study the lipid II binding as well as membrane-insertion abilities of various lantibiotics. The ultimate goal is to design novel peptide variants with superior physicochemical properties and antibacterial properties. We collaborate with Oragenics Inc., a leader in development of lantibiotics. This project is supported by a grant from Oragenics and from the NIH.
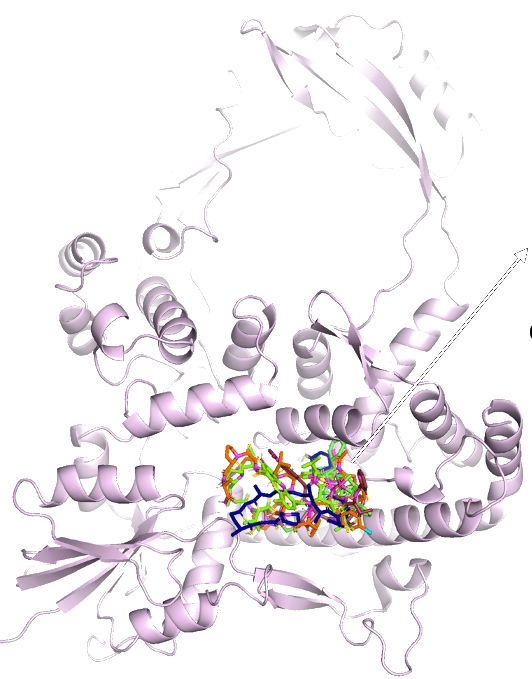
DNA/RNA interactions and in-silico drug screening of Topoisomerase IA
Topoisomerases are enzymes that relax supercoiled DNA structures formed during transcription and are novel drug targets for antibacterial and anticancer therapies. Bacterial topoisomerase I (Topo-I) remains to be exploited for antibiotics that can be used in the clinic. Inhibitors of bacterial Topo-I may provide leads for novel antibacterial drugs against pathogens resistant to current antibiotics. In collaboration with Dr. Yuk-Ching Tse Dinh, we investigate the structure and dynamics of both human and bacterial Topo-I, and use in-silico screen to identify potential Topo-I inhibitors. This project is supported by the NIH/NIGMS.